Import Mass Spectra in a mzXML Data Format
This function allows loading of spectral data files stored in a mzXML data format which in principle allows analyzing also data obtained by the VITEK® MS solution from bioMérieux.
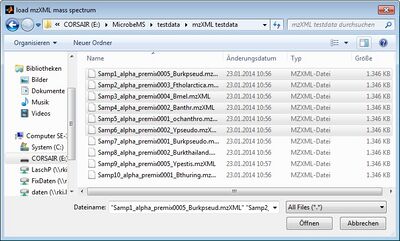
The mzXML import function of MicrobeMS is based on the Matlab routine readmzxml and is available either from the File menu bar or by the shortcut combination <Ctrl>+<X>. Select the spectral data files and press the button open to import the selected spectra.
In many cases we have noted that unprocessed mass spectra acquired by Shimadzu systems often exhibit only low signal-to-noise (S/N) levels, possible because online smooth (aka hardware smooth) has not been carried out. Therefore, it is strongly recommended to apply the Savitzky-Golay smoothing procedure with more than 30 smoothing points after data import.