Wilcoxon Rank-Sum Test: Difference between revisions
Jump to navigation
Jump to search
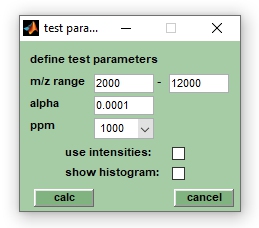
(Created page with "__FORCETOC__ == Introduction == right|input parameters for univariate t-tests| [https://en.wikipedia.org/wiki/Wilcoxon_signed-rank_test| Wilcoxon rank-sum test] (Wikipedia) To be continued == Parameter of the Wilcoxon rank-sum test == * m/z range: boundaries of the m/z region in which the Wilcoxon rank-sum tests are performed * α: significance level of the Wilcoxon rank-sum test * dx (ppm): a parameter defining the width and the number of...") |
|||
| (19 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
[[File:T-test.jpg|right|input parameters for univariate t-tests|]] | [[File:T-test.jpg|right|input parameters for univariate t-tests|]] | ||
[https://en.wikipedia.org/wiki/ | [https://en.wikipedia.org/wiki/Mann%E2%80%93Whitney_U_test| Wilcoxon rank-sum test] (Wikipedia) | ||
To be continued | To be continued | ||
| Line 18: | Line 18: | ||
== Performing Wilcoxon rank-sum tests == | == Performing Wilcoxon rank-sum tests == | ||
1. Load the mass spectral data files via the [[Load spectra (Bruker format)|load spectra]] (Bruker data file format), | 1. Load the mass spectral data files via the [[Load spectra (Bruker format)|load spectra]] (Bruker data file format), [[Import Mass Spectra in a mzXML Data Format|import spectra from mzXML data]], or the ''load MS multifile'' options of the ''File'' pulldown menu. | ||
2. Wilcoxon rank-sum tests are carried out from labeled spectra, i.e. from spectra with a [[Class Assignment|class | 2. Wilcoxon rank-sum tests are carried out from labeled spectra, i.e. from spectra with a [[Class Assignment|class assignment]]. To perform the test label two groups of spectra as class 1 and as class 2, respectively. Labeling, or class assignment, can be carried out by selecting the appropriate spectra and choosing ''class assignments'' --> ''class X'' from the ''Edit'' pulldown menu. | ||
3. The test routine always starts from original MALDI- | 3. The test routine always starts from original MALDI-ToF mass spectra, i.e. [[Spectral Pre-processing|spectral pre-processing]] and [[Peak Detection|peak detection]] is carried out automatically using pre-defined parameters. Existing pre-processed spectra and pre-defined peak tables are ignored by the Wilcoxon rank-sum tests. | ||
4. Define test parameter, such as α (significance level), the m/z range and dx (''ppm'') which has a default | 4. Define test parameter, such as α (significance level), the m/z range and dx (''ppm'') which has a default value of 1000 (relative, in ppm). The parameter dx defines the width of m/z segments in which spectra are divided during the test. Peaks found in the same m/z segment are considered identical while mass peaks in different segments are considered different peaks. | ||
5. When finished select ''peak frequency plots'' from the ''Analysis'' pulldown menu. Choose | 5. When finished select ''peak frequency plots'' from the ''Analysis'' pulldown menu. Choose options ''from selection'' or ''from class X'' if the peak frequency test should involve selected spectra or spectra with an appropriate class labeling, respectively. | ||
== Output of Wilcoxon rank-sum test == | == Output of Wilcoxon rank-sum test == | ||
| Line 46: | Line 34: | ||
{| | {| | ||
|- style="vertical-align:top;" | |- style="vertical-align:top;" | ||
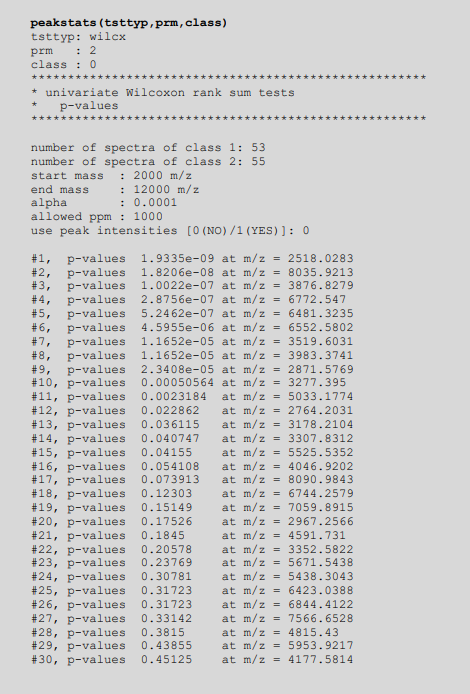
| | |[[File:Wilcox-cmdln-output.png|576px|thumb|Command line output of Wilcoxon-tests]] | ||
|[[File:Wilcox-test-plot.jpg|400px|thumb|p-values plot of the Wilcoxon-tests (log scaled): The smaller the p-value at the specific m/z position the higher the discriminative potential of biomarker peaks at this position]] | |||
|[[File:Wilcox-test-plot.jpg|400px|thumb|p-values plot of the Wilcoxon-tests (log scaled) | |||
|} | |} | ||
Latest revision as of 17:23, 11 April 2025
Introduction
Wilcoxon rank-sum test (Wikipedia)
To be continued
Parameter of the Wilcoxon rank-sum test
- m/z range: boundaries of the m/z region in which the Wilcoxon rank-sum tests are performed
- α: significance level of the Wilcoxon rank-sum test
- dx (ppm): a parameter defining the width and the number of the m/z spectra segments. For example, a spectral segment centered at the position x covers a m/z interval of the width x * dx/10^6. The boundaries of the spectra segments are defined by [x*(1-dx/(2*10^6))] and [x*(1+dx/(2*10^6))], respectively. For values of x = 2000 (m/z) and dx = 1000, the width of the respectice segment is 2 and the m/z values of the boundaries are 1999 and 2001.
- intensity: defines if barcode spectra or peak weighting factors are utilized as test inputs.
- show histogram: provides a histogram of the test outputs (p-values, AUC, etc.) and gives also the mean, median and the standard deviation of the test variables.
Performing Wilcoxon rank-sum tests
1. Load the mass spectral data files via the load spectra (Bruker data file format), import spectra from mzXML data, or the load MS multifile options of the File pulldown menu.
2. Wilcoxon rank-sum tests are carried out from labeled spectra, i.e. from spectra with a class assignment. To perform the test label two groups of spectra as class 1 and as class 2, respectively. Labeling, or class assignment, can be carried out by selecting the appropriate spectra and choosing class assignments --> class X from the Edit pulldown menu.
3. The test routine always starts from original MALDI-ToF mass spectra, i.e. spectral pre-processing and peak detection is carried out automatically using pre-defined parameters. Existing pre-processed spectra and pre-defined peak tables are ignored by the Wilcoxon rank-sum tests.
4. Define test parameter, such as α (significance level), the m/z range and dx (ppm) which has a default value of 1000 (relative, in ppm). The parameter dx defines the width of m/z segments in which spectra are divided during the test. Peaks found in the same m/z segment are considered identical while mass peaks in different segments are considered different peaks.
5. When finished select peak frequency plots from the Analysis pulldown menu. Choose options from selection or from class X if the peak frequency test should involve selected spectra or spectra with an appropriate class labeling, respectively.
Output of Wilcoxon rank-sum test
Example of the output from a series of Wilcoxon rank-sum tests taken from the log file of MicrobeMS: