Peak Frequency Test: Difference between revisions
(Created page with "== Introduction == Peak frequency tests are useful for biomarker screening in ensembles of mass spectra exhibiting certain degree of similarity. These tests require pre-processed spectra with valid peak tables as inputs and are suitable to systematically determine the number of peaks and their relative abundance throughout the spectral m/z intervals. For peak frequency tests it is required to compile a mass spectral data set and to perform pre-processing including peak...") |
|||
| Line 5: | Line 5: | ||
== Performing a peak frequency test == | == Performing a peak frequency test == | ||
1. Load the mass spectral data files via the [[Load spectra (Bruker format)|load spectra]] (Bruker data file format), | 1. Load the mass spectral data files via the [[Load spectra (Bruker format)|load spectra]] (Bruker data file format), [[Import Mass Spectra in a mzXML Data Format|import spectra from mzXML data]], or the ''load MS multifile'' options of the ''File'' pulldown menu. | ||
2. Perform [[Spectral Pre-processing|spectral pre-processing]] and [[Peak Detection|peak detection]]. | 2. Perform [[Spectral Pre-processing|spectral pre-processing]] and [[Peak Detection|peak detection]]. | ||
3. Note that spectra selected for peak frequency tests are required to contain valid peak tables. | 3. Note that spectra selected for peak frequency tests are required to contain valid peak tables. Spectra without associated peak table cannot be processed. | ||
4. Select the respective mass spectra in the listbox at the top left corner (the listbox is labeled by | 4. Select the respective mass spectra in the listbox at the top left corner (the listbox is labeled by ''MicrobeMS spectra ID`s''). To select multiple spectra hold the <shift> key while selecting. Alternatively, frequency tests can be also carried out from labeled spectra, i.e. from spectra with a [[Class Assignment|class assignment]]. Labeling, or class assignment, can be carried out by selecting the appropriate spectra and choosing ''class assignments'' --> ''class X'' from the ''Edit'' pulldown menu. | ||
5. If necessary define the parameter dx (''ppm tolerance'') which has a default value of dx = 1000 | 5. If necessary define the parameter dx (''ppm tolerance'') which has a default value of dx = 1000 (relative ppm units). The parameter dx defines the width of m/z segments in which spectra are divided when analyzing peak frequencies. Peaks found in the same m/z segment are considered identical while mass peaks in different segments are considered different peaks. An spectral segment at the position x has width of x * dx/10^6 and ranges from [x*(1-dx/(2*10^6))] to [x*(1+dx/(2*10^6))], for example from m/z 1999 to 2001 if x = 2000 (m/z) and dx = 1000. | ||
6. When finished select ''peak frequency plots'' from the ''Analysis'' pulldown menu. Choose | 6. When finished select ''peak frequency plots'' from the ''Analysis'' pulldown menu. Choose options ''from selection'' or ''from class X'' if the peak frequency test should involve selected spectra or spectra with an appropriate class labeling, respectively. | ||
== Output of a peak frequency test == | == Output of a peak frequency test == | ||
Revision as of 13:53, 16 December 2024
Introduction
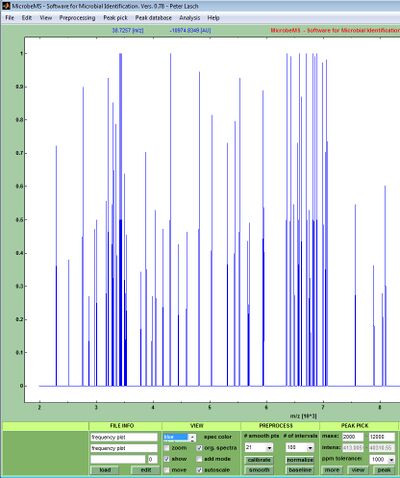
Peak frequency tests are useful for biomarker screening in ensembles of mass spectra exhibiting certain degree of similarity. These tests require pre-processed spectra with valid peak tables as inputs and are suitable to systematically determine the number of peaks and their relative abundance throughout the spectral m/z intervals. For peak frequency tests it is required to compile a mass spectral data set and to perform pre-processing including peak picking using standardized parameters. Peak frequency tests include a systematic evaluation for peak abundance in specific m/z segments which width is defined by a program-wide parameter dx (sometimes denoted as ppm tolerance). The output of a peak frequency test is a table in which ranked absolute and relative peak frequency values are listed. In addition, the relative peak frequency is plotted for each m/z segment (see screenshot below).
Performing a peak frequency test
1. Load the mass spectral data files via the load spectra (Bruker data file format), import spectra from mzXML data, or the load MS multifile options of the File pulldown menu.
2. Perform spectral pre-processing and peak detection.
3. Note that spectra selected for peak frequency tests are required to contain valid peak tables. Spectra without associated peak table cannot be processed.
4. Select the respective mass spectra in the listbox at the top left corner (the listbox is labeled by MicrobeMS spectra ID`s). To select multiple spectra hold the <shift> key while selecting. Alternatively, frequency tests can be also carried out from labeled spectra, i.e. from spectra with a class assignment. Labeling, or class assignment, can be carried out by selecting the appropriate spectra and choosing class assignments --> class X from the Edit pulldown menu.
5. If necessary define the parameter dx (ppm tolerance) which has a default value of dx = 1000 (relative ppm units). The parameter dx defines the width of m/z segments in which spectra are divided when analyzing peak frequencies. Peaks found in the same m/z segment are considered identical while mass peaks in different segments are considered different peaks. An spectral segment at the position x has width of x * dx/10^6 and ranges from [x*(1-dx/(2*10^6))] to [x*(1+dx/(2*10^6))], for example from m/z 1999 to 2001 if x = 2000 (m/z) and dx = 1000.
6. When finished select peak frequency plots from the Analysis pulldown menu. Choose options from selection or from class X if the peak frequency test should involve selected spectra or spectra with an appropriate class labeling, respectively.
Output of a peak frequency test
Example of a peak frequency table taken from the log file of MicrobeMS:
|
peakstats(tsttyp,prm,class) tsttyp: frequ prm : 0 class : 1 * peak frequency charts ****************************** * peak frequencies from class 1 * 108 - number of spectra * 1000 - ppm ****************************************************** 1 - rel frequency: 100% (108 absolute) at m/z 6888.5144 2 - rel frequency: 100% (108 absolute) at m/z 6817.3196 3 - rel frequency: 100% (108 absolute) at m/z 6702.1082 4 - rel frequency: 100% (108 absolute) at m/z 6572.99 5 - rel frequency: 100% (108 absolute) at m/z 6354.3125 6 - rel frequency: 100% (108 absolute) at m/z 4306.6995 7 - rel frequency: 100% (108 absolute) at m/z 3445.7725 8 - rel frequency: 100% (108 absolute) at m/z 3423.7905 9 - rel frequency: 100% (108 absolute) at m/z 3410.2222 10 - rel frequency: 99.0741% (107 absolute) at m/z 6844.4122 11 - rel frequency: 99.0741% (107 absolute) at m/z 6423.0388 12 - rel frequency: 98.1481% (106 absolute) at m/z 7059.8915 13 - rel frequency: 97.2222% (105 absolute) at m/z 6989.2526 14 - rel frequency: 94.4444% (102 absolute) at m/z 4815.43 15 - rel frequency: 92.5926% (100 absolute) at m/z 5525.5352 16 - rel frequency: 92.5926% (100 absolute) at m/z 3211.5771 17 - rel frequency: 89.8148% (97 absolute) at m/z 2764.2031 18 - rel frequency: 88.8889% (96 absolute) at m/z 5932.7762 19 - rel frequency: 87.037% (94 absolute) at m/z 6612.9588 20 - rel frequency: 85.1852% (92 absolute) at m/z 3287.9415 21 - rel frequency: 83.3333% (90 absolute) at m/z 9626.3733 22 - rel frequency: 81.4815% (88 absolute) at m/z 5033.1774 23 - rel frequency: 79.6296% (86 absolute) at m/z 5438.3043 24 - rel frequency: 78.7037% (85 absolute) at m/z 3352.5822 25 - rel frequency: 73.1481% (79 absolute) at m/z 6552.5802 26 - rel frequency: 73.1481% (79 absolute) at m/z 5303.6301 27 - rel frequency: 72.2222% (78 absolute) at m/z 2297.6308 28 - rel frequency: 70.3704% (76 absolute) at m/z 7032.9167 29 - rel frequency: 70.3704% (76 absolute) at m/z 3876.8279 30 - rel frequency: 64.8148% (70 absolute) at m/z 3307.8312
|